
Microsoft Unveils Biomolecular Emulator-1 (BioEmu-1): A Game Changer in Protein Motion Analysis and Drug Discovery.
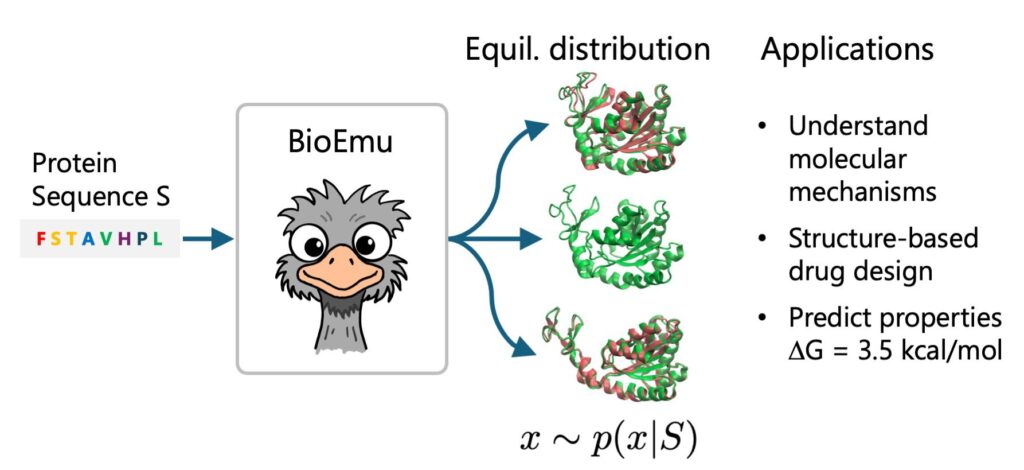
Microsoft has made a groundbreaking leap in biotechnology with the launch of its AI system, Biomolecular Emulator-1 (BioEmu-1), a deep learning model designed to simulate protein motion at unprecedented speeds. Capable of generating thousands of protein structures per hour on a single GPU, BioEmu-1 is set to revolutionize drug discovery and molecular biology, offering researchers powerful tools to understand disease pathways and design better therapeutic agents faster.
What is Biomolecular Emulator-1 (BioEmu-1)?
Biomolecular Emulator-1 (BioEmu-1) is Microsoft’s latest venture into the field of AI driven biotechnology. Developed in collaboration with leading computational biologists and powered by Azure cloud infrastructure, BioEmu-1 is an advanced deep learning system designed to model the movements and structures of proteins in three dimensional space.
Protein motion is a fundamental process in biology, dictating how molecules interact, signal, and function within living organisms. Traditional methods like molecular dynamics (MD) simulations are computationally intensive and time consuming. BioEmu-1 bypasses these limitations by using deep generative models that mimic protein behavior in real time.
With BioEmu-1, Microsoft is not only contributing to faster biomedical research but also positioning itself as a major player in AI powered life sciences.
Why Protein Motion Matters in Drug Discovery
Proteins are molecular machines of life. Their shape, flexibility, and dynamic motion govern everything from enzyme activity to immune system response. Understanding how a protein moves or changes shape can reveal hidden drug binding sites and potential therapeutic targets.
Unfortunately, capturing protein motion is incredibly challenging:
- Experimental techniques like cryo-electron microscopy (Cryo-EM) provide static snapshots.
- Molecular simulations are precise but computationally heavy.
This is where Biomolecular Emulator-1 (BioEmu-1) becomes revolutionary. By generating thousands of possible protein conformations per hour, researchers can now model real time dynamics and predict how proteins behave under different biological conditions.
How Does Biomolecular Emulator-1 (BioEmu-1) Work?
Microsoft’s Biomolecular Emulator-1 (BioEmu-1) uses deep generative models a type of machine learning that learns from existing protein motion data and generates plausible future structures.
Core Features:
- Single GPU Efficiency: Generates protein conformations on a single graphics processing unit (GPU), making it cost effective and scalable.
- High Throughput: Capable of producing thousands of conformations per hour, ideal for mass screening in drug research.
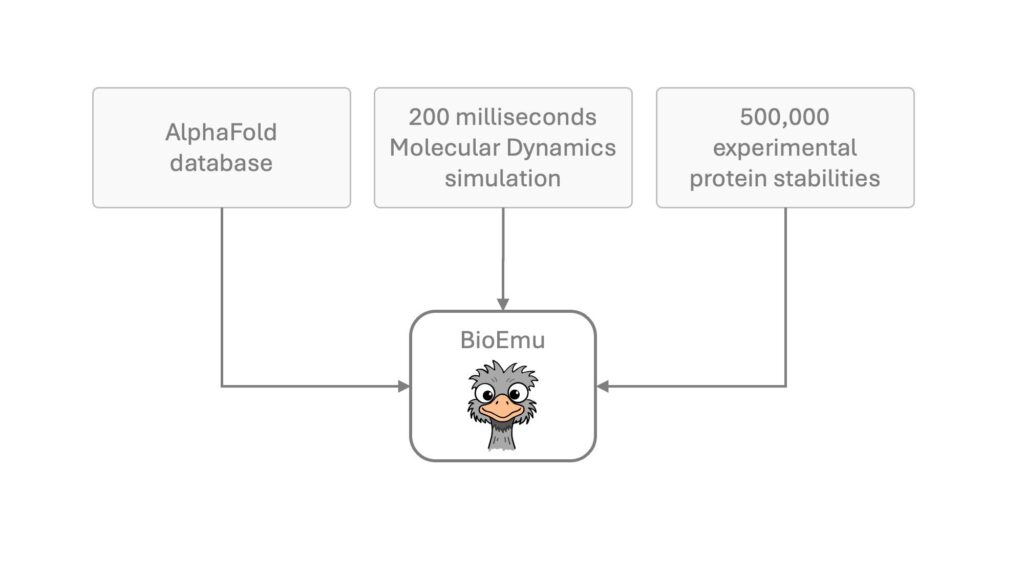
- Data Driven Accuracy: Trained on large protein motion datasets, ensuring scientific reliability.
By simulating realistic trajectories of protein folding and unfolding, BioEmu-1 enables drug designers to predict which conformation will interact best with potential drug molecules a vital step in structure based drug design.
Applications of Biomolecular Emulator-1 (BioEmu-1)
1. Accelerated Drug Discovery
The most transformative impact of Biomolecular Emulator-1 (BioEmu-1) is in pharmaceutical research. Instead of spending months on laboratory experiments or expensive simulations, scientists can now:
- Rapidly test drug-protein interactions.
- Explore rare or transient protein conformations.
- Optimize drug candidates before clinical trials.
This can reduce drug discovery timelines by up to 50%, saving millions in research costs.
2. Understanding Rare Diseases
For genetic diseases involving protein misfolding (e.g., cystic fibrosis, Huntington’s), BioEmu-1 offers insights into abnormal protein motion, potentially identifying novel treatment avenues.
3. Vaccine and Antibody Design
During the COVID-19 pandemic, researchers learned the importance of understanding viral protein behavior. Biomolecular Emulator-1 (BioEmu-1) can simulate how viral proteins like spike proteins evolve, helping scientists design more effective vaccines and antibodies.
How BioEmu-1 Stands Out from Other AI Models
There are other models like AlphaFold by DeepMind, which predict static protein structures. However:
| Feature | AlphaFold | Biomolecular Emulator-1 |
|---|---|---|
| Focus | Static protein structure | Dynamic protein motion |
| Output | One conformation | Thousands/hour |
| Use Case | Structure prediction | Drug design, protein dynamics |
| Efficiency | High resource use | Single-GPU capable |
This makes Biomolecular Emulator-1 (BioEmu-1) not a competitor, but a complement to models like AlphaFold, expanding the toolkit available for computational biology.
Deep Learning Behind BioEmu-1
BioEmu-1 is built using a type of AI called generative neural networks. These networks:
- Learn from time-series data of protein movements.
- Understand patterns of flexibility, folding, and transformation.
- Generate new, valid conformations that fit biological rules.
Its architecture likely uses transformers and diffusion models, popular in recent generative AI systems (like text and image generation), adapted for molecular 3D data.
Microsoft also incorporated physics informed neural networks (PINNs) to ensure that the outputs remain biologically and chemically plausible.
Microsoft’s Vision for AI in Life Sciences
By launching Biomolecular Emulator-1 (BioEmu-1), Microsoft demonstrates its growing commitment to AI for health, life sciences, and precision medicine.
Satya Nadella, Microsoft’s CEO, has consistently emphasized AI’s potential in solving humanity’s biggest challenges. In this context, BioEmu-1 aligns perfectly with Microsoft’s AI for Good initiative, targeting real-world applications beyond enterprise software.
The system is also integrated with Azure Quantum Elements, indicating Microsoft’s intent to unify AI, quantum computing, and molecular biology in a single research ecosystem.
Integration with Microsoft Cloud & Research Ecosystem
Microsoft is ensuring that Biomolecular Emulator-1 (BioEmu-1) is accessible via the Azure platform, so that researchers, pharmaceutical companies, and academic labs can:
- Run simulations using Microsoft’s cloud infrastructure.
- Integrate protein models into existing bioinformatics pipelines.
- Leverage Microsoft Copilot for automatic analysis and reporting.
Additionally, Azure OpenAI integration allows users to query results in natural language, making BioEmu-1 accessible even to non-specialists in computational biology.
Real World Impact and Collaborations
Microsoft has already begun collaborating with major pharmaceutical firms, research universities, and medical institutes to deploy Biomolecular Emulator-1 (BioEmu-1) in active research settings.
Examples include:
- Cancer therapeutics: Modeling protein targets involved in tumor progression.
- Neurodegenerative disorders: Understanding misfolded protein pathways in Alzheimer’s.
- Infectious diseases: Tracking viral protein mutations to improve vaccine effectiveness.
Early feedback suggests that BioEmu-1 significantly improves lead identification and pre-clinical testing accuracy.
Challenges and Ethical Considerations
Despite its potential, Biomolecular Emulator-1 (BioEmu-1) also raises key challenges:
Bias in training data: Like any AI, it is only as good as the data it learns from.
Misuse of biological simulations: Potential misuse in creating synthetic pathogens.
Access and equity: Ensuring that smaller labs and developing nations can use the system affordably.
Microsoft says it is committed to open science principles, and may consider offering BioEmu-1 as an open source tool for academic use in the future.
What’s Next for BioEmu-1?
Microsoft has laid out ambitious future plans for Biomolecular Emulator-1 (BioEmu-1):
- Adding support for RNA and DNA modeling.
- Integrating with quantum simulation tools for hybrid modeling.
- Expanding into plant biology and environmental biochemistry.
The next versions of BioEmu-1 may also feature multi-scale modeling, combining protein dynamics with cell level simulations to understand how molecular events impact larger biological systems.
Conclusion
Biomolecular Emulator-1 (BioEmu-1) is a powerful testament to what happens when cutting edge AI meets the frontiers of molecular biology. It offers unprecedented speed, scalability, and accuracy in modeling protein dynamics crucial for accelerating drug discovery and understanding life at a fundamental level.
As Microsoft continues to develop this technology, BioEmu-1 could become a cornerstone tool in labs around the world, reshaping how we approach medicine, disease, and even evolution itself.
With its ability to generate thousands of realistic protein structures per hour, and run on accessible computing hardware, Biomolecular Emulator-1 (BioEmu-1) is not just an innovation it’s a scientific revolution in motion.
Stay Connected for more updates on artificial intelligence, drug discovery, and innovations in biotechnology, follow The News Drill.
💬 Send your tips or stories to: editor@thenewsdrill.com
📩 General inquiries: contact@thenewsdrill.com
FAQs
What is Biomolecular Emulator-1 (BioEmu-1)?
Biomolecular Emulator-1 (BioEmu-1) is an AI system by Microsoft that simulates protein motion using deep learning. It generates thousands of protein conformations per hour on a single GPU, transforming drug discovery and molecular biology.
How does BioEmu-1 differ from AlphaFold?
AlphaFold predicts static protein structures, while BioEmu-1 focuses on dynamic protein motion. BioEmu-1 simulates how proteins behave and evolve, making it more useful for drug design and real-time biological modeling.
What are the key applications of BioEmu-1?
BioEmu-1 is used in accelerated drug discovery, protein misfolding studies, rare disease research, and the development of vaccines and antibodies.
Can BioEmu-1 be used by pharmaceutical companies?
Yes, BioEmu-1 is designed for integration with cloud platforms like Azure, making it highly accessible and scalable for pharmaceutical research and biotech organizations.
How fast is BioEmu-1 compared to traditional methods?
BioEmu-1 can simulate thousands of protein conformations per hour using just a single GPU, which is significantly faster than traditional molecular dynamics simulations that require days of supercomputing time.
Is BioEmu-1 available for public or academic use?
Currently, Microsoft is collaborating with selected research partners. Broader academic access is expected to be offered through Azure in future phases.
How does BioEmu-1 support drug discovery?
By modeling thousands of dynamic protein structures, BioEmu-1 helps researchers find the best binding conformations for drug molecules, accelerating lead identification and preclinical validation.
Is Biomolecular Emulator-1 open-source?
No, BioEmu-1 is not currently open-source. However, Microsoft may expand access to academic institutions and researchers under certain partnerships or licensing models.
Does BioEmu-1 require powerful hardware?
No. One of the major advantages of BioEmu-1 is that it runs on a single GPU, making it affordable and accessible without requiring high-performance supercomputers.
Where can I learn more about BioEmu-1?
You can follow Microsoft Research, visit Azure Quantum Elements, or stay updated with technology insights at The News Drill for more on BioEmu-1 and its developments.













